Our Services:
Computational RNA-Focused Modeling & Drug Discovery
At Darien Biodiscovery, we specialize in cutting-edge computational solutions to unlock the therapeutic potential of RNA. As a virtual Contract Research Organization (CRO), we combine advanced algorithms, AI/ML-driven workflows, and deep expertise in RNA biology to deliver tailored services for academic researchers, biotech innovators, and pharmaceutical partners. Below is an overview of our core offerings:
RNA-Ligand Interaction Modeling
Virtual Screening & Docking:
Screen small-molecule libraries against RNA targets (e.g., riboswitches, viral RNA elements, human lncRNAs) to identify high-affinity binders.
Optimize lead compounds using deep learning-based scoring functions, end-point scoring methods, and RBFE calculations.
G-Quadruplex (G4) Targeting:
Design small molecules to stabilize or disrupt oncogenic/neurodegenerative RNA G-quadruplexes.
Validate binding modes with MD simulations and in silico mutagenesis.
Fragment-Based Drug Discovery (FBDD):
Identify fragment hits targeting RNA pockets and evolve them into drug-like molecules.
RNA Structure Prediction & Modeling
Secondary & Tertiary Structure Prediction:
Accurately model RNA secondary structures (helices, loops, bulges) and 3D conformations using cutting-edge deep learning and physics-based tools.
Identify functional motifs (e.g., G-quadruplexes, pseudoknots) critical for RNA-ligand interactions.
Dynamic RNA Folding Analysis:
Predict co-transcriptional folding pathways and transient structural states for mechanistic insights.
RNA-Protein Complex Modeling:
Characterize RNA-protein binding interfaces to guide interventions in regulatory networks.


RNA-Focused Virtual Chemical Library Design & Navigation
RNA-Targeted Drug Discovery Platforms
RNA-Targeted Chemical Space Design
Custom Library Construction:
Our team builds RNA-optimized libraries by applying physicochemical filters (electrostatic complementarity, hydrogen-bonding patterns) and structural motifs (G-quadruplex groove binders, loop-targeting scaffolds).Privileged Motif Integration:
We enrich libraries with RNA-binding chemotypes (e.g., aminoglycoside-inspired cores, polycationic linkers) to boost hit rates.
Gigascale Library Generation & Curation
>10 Billion Compound Access:
We generate non-enumerated chemical spaces on-demand using proprietary rules (SMIRKS/SMARTS), genetic algorithms, and generative graph models – no client storage/computational burden.On-the-Fly Virtual Synthesis:
Our workflows dynamically assemble molecules from building blocks, exploring vast regions of chemical space without pre-enumeration.
Generative AI-Driven Library Expansion
De Novo Molecule Design:
Using deep learning models (VAEs, GANs) trained on proprietary RNA-ligand datasets, we generate novel, synthetically feasible compounds with high RNA-binding potential.Reinforcement Learning (RL) Optimization:
We iteratively refine libraries against your target’s RNA-specific objectives (e.g., binding affinity, selectivity, metabolic stability).
Ultra-Large Virtual Screening (ULVS)
GPU-Accelerated Docking:
We screen gigascale libraries against your RNA target using optimized docking pipelines (AutoDock-GPU, DiffDock) and ML surrogate models.Pharmacophore-Guided Prioritization:
We focus computational resources on RNA "hotspots" (e.g., G-quadruplex grooves, pseudoknot interfaces) to accelerate hit identification.
5. Non-Enumerated Space Navigation
Fragment/Reaction-Driven Exploration:
We traverse ultra-large chemical spaces using fragment growing, morphing, and retrosynthetic rules to identify novel RNA binders.Chemical Space Mapping:
Using topological data analysis (TDA), we visualize RNA-targeted "islands" in chemical space and prioritize regions for synthesis.


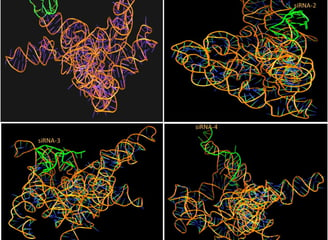
Structure-Based Drug Design (SBDD)
Rational Inhibitor/Modulator Design:
We leverage RNA 3D structures (experimental or predicted) to design small molecules or oligonucleotides with improved specificity and binding affinity.Binding Mode Refinement:
Our team performs binding free energy calculations (MM/PBSA, FEP) and MD simulations to optimize interactions with your RNA target.
Machine Learning-Driven Optimization
Custom Predictive Models:
We train ML models on your data (or our proprietary RNA datasets) to predict RNA-ligand binding, ADMET properties, and off-target risks.Lead Optimization Roadmaps:
Receive prioritized chemical modifications and scaffold-hopping strategies to enhance efficacy and safety.
CRISPR RNA (crRNA/gRNA) Design
Off-Target Minimization:
We computationally design guide RNAs for CRISPR-Cas systems with improved on-target specificity using RNA:DNA mismatch penalty scoring and genome-wide homology screening.Delivery-Optimized Sequences:
We engineer chemical modification patterns (e.g., 2’-O-methyl, phosphorothioate) to enhance gRNA stability in silico.
siRNA & ASO Optimization
Target Accessibility Prediction:
We predict RNA secondary structure to identify accessible regions for siRNA/ASO binding.Chemical Modification Blueprints:
Receive customized modification patterns (LNA, 2’-MOE, GalNAc conjugates) to optimize stability, potency, and tissue targeting.
Specialized Services
RNA Chemical Modification Analysis:
Stability & Function Prediction: We model how modifications (e.g., m6A, pseudouridine, 5-methylcytidine) impact RNA structure, protein binding, and degradation resistance.
RNA-Protein Interaction Networks:
Interactome Mapping: We predict RNA-protein binding partners using sequence/structure motifs and prioritize druggable hubs for your disease target.
Data Integration & Visualization:
Publication-Ready Outputs: Receive high-quality 3D renderings, interaction heatmaps, and kinetic simulation videos – no software installation required.


Deliverables
Ready-to-Test Hit Lists:
Ranked small molecules with predicted binding modes, synthesis routes or supplier codes, and ADMET profiles.
Strategic Reports:
Chemical space maps, SAR insights, and optimization roadmaps.
Results instead of platform:
We execute workflows using our proprietary and FOSS tools – clients receive results, not software platform, thus saving the time.
Why This Model Works for Clients
Zero Infrastructure Overhead:
We handle all computational costs (GPU clusters, software licenses, data storage).
RNA-Specific Focus:
Libraries pre-filtered for RNA compatibility, avoiding wasted resources on protein-centric chemistries.
IP Ownership:
You retain full rights to all designed molecules and screening data.
Innovative Solutions
We Turn Visionary Concepts into Actionable Results – No Client Software, Only Expert Execution


RNA-Targeted Generative Chemistry
How We Deliver:
Our team combines generative AI, quantum-inspired algorithms, and evolutionary chemistry to explore RNA-binding chemical spaces inaccessible to traditional methods.
Your Results:
Novel chemotype designs against “undruggable” RNA targets
Synthesizable compound libraries with prioritized synthesis routes

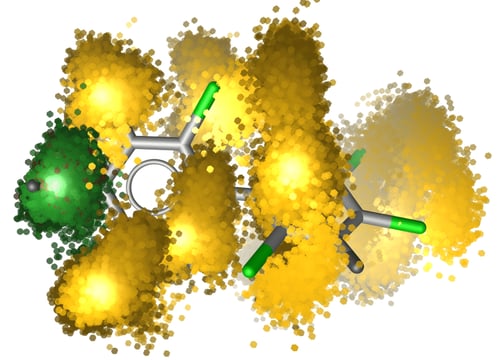

Dynamic Structure-Activity Relationships (DSAR)
How We Deliver:
We perform adaptive sampling MD and Markov state modeling to capture RNA-ligand interaction dynamics across microsecond timescales.
Your Results:
Allosteric modulator candidates with kinetic binding profiles
Transient pocket discovery reports for patent strategy
RNA Editing Design Service
How We Deliver:
We provide end-to-end computational design for RNA base editing systems:
Off-target-minimized gRNA sequences
Editor protein-RNA interface optimization
Your Results:
Ready-to-test gRNA designs with off-target risk assessment reports
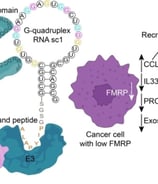
RNA-PROTAC Co-Development
How We Deliver:
We execute full computational design of RNA-targeting PROTACs:
E3 ligase warhead pairing analysis
Linker-length optimization for ternary complex stability
Your Results:
PROTAC candidate designs with degradation efficiency predictions
RNA:Small Molecule Combination Therapy
How We Deliver:
We design synergistic RNA-targeted combinations via:
siRNA/ASO + small molecule joint optimization
Resistance mechanism evasion modeling
Your Results:
Validated combination protocols with dosing schedules
“Our lab had spent two years trying to target a conserved RNA element in enterovirus without success. Larger CROs dismissed the project as ‘too niche,’ but Darien’s team embraced the challenge. Despite their small size, they delivered a prioritized hit list within 6 weeks using their generative chemistry platform. What impressed me most was their responsiveness – Dr. Laar, their CEO, was ready to join the calls to troubleshoot, something unheard of with bigger providers. The lead compound they identified is now central to our ongoing grant proposal. For specialized RNA projects, size is an advantage: Darien moves faster, cares more, and costs less.”
Prof Li W., Head of Lab, Shanghai, China
Antiviral RNA Target Breakthrough
