At Darien Biodiscovery, we have built a proprietary technology stack specifically engineered to tackle RNA’s unique challenges – from its dynamic structures to its elusive binding pockets. Our hybrid approach combines physics-based rigor with AI’s pattern recognition power, enabling us to deliver results where traditional methods fail.
Core Technology Pillars
RNA Spatial Structure Modeling
a. Physics-Based Foundations
All-Atom Molecular Dynamics (MD): Simulate RNA folding & ligand binding at μs-ms timescales using specialized force fields (RNA-OL3, AMBER-DH2).
Enhanced Sampling: Accelerate rare-event sampling with GaMD, MetaDynamics, and AI-driven collective variable discovery.
Co-Transcriptional Folding Predictions: Model RNA structure formation in real-time as nucleotides are synthesized.
b. Deep Learning Revolution
3D Capsule Networks: Predict tertiary structures from sequence alone using our DeepFoldRNA™ model, trained on 1,500+ experimentally solved RNA structures.
Ensemble Modeling: Generate probabilistic structure ensembles to capture RNA’s intrinsic flexibility.
RNA-Focused Virtual Screening
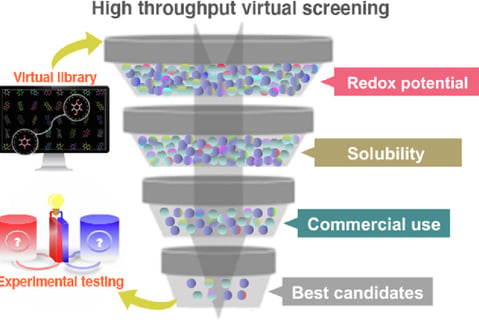
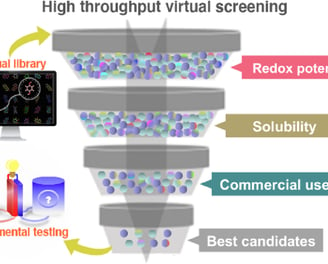
a. Gigascale Docking Infrastructure
GPU-Accelerated Workflows: Screen 10B+ compounds in days using optimized AutoDock-GPU pipelines.
Target-Adaptive Grids: Dynamically adjust docking grids to RNA’s flexible regions (loops, bulges).
b. Machine Learning Filters
RNA-ChemSpace Navigator™: Prune non-RNA-compatible chemotypes using 15+ RNA-specific filters (charge, solvation, motif matching).
Non-Enumerated Library Support: Screen ultra-large spaces via SMIRKS-based reaction rules & generative subgraph sampling.
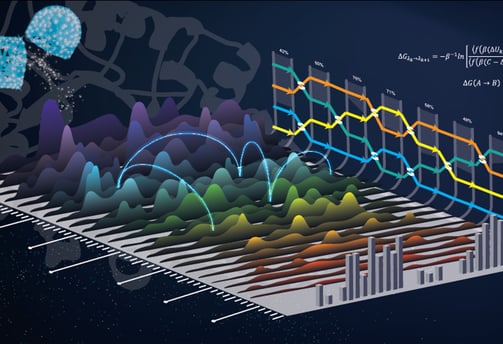
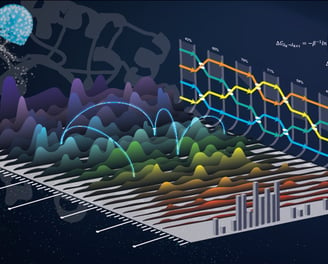
Lead Optimization Engine
a. Free Energy Mastery
Alchemical FEP: Calculate ΔΔG binding with <1 kcal/mol error using bespoke RNA solvent models.
QM/MM Hybrid: Refine critical interactions (e.g., cation-π, halogen bonds) at the DFT level.
b. AI-Driven Property Prediction
ADMET-RNA™: Predict tissue penetration, efflux ratios, and RNA-specific toxicity risks.
Synthetic Accessibility Scoring: Prioritize compounds with 3-click MEDCHEM routes using our retrosynthesis-trained AI.
Proprietary Breakthrough: RiboAffinity
Our key technology pillar – the first AI scoring function trained exclusively on RNA-ligand dynamic interactions – redefines affinity prediction accuracy:
Why It’s State-of-the-Art:
Largest Training Set: >250 experimental nucleic acid-ligand complexes augmented with template-based modeling and molecular dynamics data and binding data points (Kd, IC50, ΔG) across various RNA target classes.
Multimodal Input: Processes 3D RNA-ligand poses, sequence context, and solvent dynamics.
Outperforms Legacy Tools: 30% higher enrichment vs. Glide/AutoDock in our in-house RNA benchmark sets.
Key Applications:
Rank-order virtual screening hits with experimental-level accuracy
Explain binding via attention maps, highlighting critical RNA nucleotides
Technology Integrations
Generative Chemistry: VAEs & diffusion models for RNA-targeted library expansion.
CRISPR Design Suite: Optimize guide RNAs via in-house deep learning models (DeepGuide™).
Cloud-Native Architecture: AWS-optimized pipelines for burst scaling to 128+ GPUs.
Why Our Technology Wins
Hybrid AI/Physics Approach: Get the robustness of physics with AI’s speed.
RNA-Specific Tuning: No more repurposed protein tools.
Scalability Meets Precision: From gigascale screens to quantum-level refinements.
